
China Animal Husbandry & Veterinary Medicine ›› 2026, Vol. 53 ›› Issue (2): 819-836.doi: 10.16431/j.cnki.1671-7236.2026.02.028
• Genetics and Breeding • Previous Articles Next Articles
DU Zhiwen1( ), GAO Yuxin1, LIU Shuqin1, MA Haibin2, YANG Lei1, SONG Lishuang1, BAI Chunling1, WEI Zhuying1, LI Guangpeng1(
), GAO Yuxin1, LIU Shuqin1, MA Haibin2, YANG Lei1, SONG Lishuang1, BAI Chunling1, WEI Zhuying1, LI Guangpeng1( ), SU Guanghua1(
), SU Guanghua1( )
)
Received:2025-05-27
Online:2026-02-20
Published:2026-01-27
Contact:
LI Guangpeng, SU Guanghua
E-mail:dzw15133339850@163.com;gpengli@imu.edu.cn;guanghuasu@imu.edu.cn
CLC Number:
DU Zhiwen, GAO Yuxin, LIU Shuqin, MA Haibin, YANG Lei, SONG Lishuang, BAI Chunling, WEI Zhuying, LI Guangpeng, SU Guanghua. Efficiency Optimization of MSTN Gene Editing in Huaxi Cattle Cells and Embryos[J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(2): 819-836.
Table 1
Primers for MSTN gene"
引物 Primers | 正向序列 Forward sequences (5′→3′) | 反向序列 Reverse sequences (5′→3′) | 片段长度 Fragment length/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| W1 | ACGTTTGGCTTGGCGTTACT | CACCAGCAGGACTACTCACA | 479 | 55 |
| W2 | GGAGGTGTTCGTTCATTTTTCA | CTGGGGTAAGGTGCCTTTGT | 566 | 53 |
| W3 | GCCATAAAGGAAGAATCAAGCC | GACCTTCCATGTTTGAGGAAGC | 592 | 53 |
Table 2
sgRNA sequences for MSTN gene"
| sgRNAs | 正向序列 Forward sequences (5′→3′) | 反向序列 Reverse sequences (5′→3′) |
|---|---|---|
| sgRNA1-1 | CACCAAGACGATGACTACCACGCC | AAACGGCGTGGTAGTCATCGTCTT |
| sgRNA1-2 | CACCCGATGACTACCACGCCAGGA | AAACTCCTGGCGTGGTAGTCATCG |
| sgRNA1-3 | CACCTGACCGTTTCCGTCCTGGCG | AAACCGCCAGGACGGAAACGGTCA |
| sgRNA2-1 | CACCATGGGTTTGATGAGTCTC | AAACGAGACTCATCAAACCCAT |
| sgRNA2-2 | CACCAGAACCAGGAGAAGATGGAC | AAACGTCCATCTTCTCCTGGTTCT |
| sgRNA2-3 | CACCACTGTCGCAGGAGTCTTGAC | AAACGTCAAGACTCCTGCGACAGT |
Table 3
Primers used for off-target site detection"
脱靶位点 Off-target sites | 正向序列 Forward sequences (5′→3′) | 反向序列 Reverse sequences (5′→3′) |
|---|---|---|
| OF1 | GCTCACCTGTCCCCTAGTTTT | AACTTCCAGATTGCTGACCG |
| OF2 | AGTGAAGAAGCAATAACTGGTCA | GGCAGACAGACATGAAGGCT |
| OF3 | CCAGCAGACAGCTAACAGTTC | ACAGTCATGGTCACTGATGGAG |
| OF4 | TTCCAGACCTACTGAGGGAGG | AAGAAGAGACGGTGTTGCTGA |
| OF5 | CACCTGTCAGTTGACTACACT | TGTGCTTAGTCATCTTCCCAGA |
| OF6 | CACTCTAGGGAACCAACGGG | ATTTTGTGCCTCCGCTCACT |
| OF7 | CCTCCTGCCACACCTTTATCA | AGCAGCAAAGGTTTACGGGG |
| OF8 | GGTTCTGTTCCTTACGGGGT | TCCTGTCGTTTGTCCTTGCT |
| OF9 | CTGGCCACTCTGATTCGGTA | GGTACAAACCCCAGCCAGTT |
| OF10 | TCTGGGGAATCGGGTTACCT | ATTTTCCCTGTGGAGGTGGC |
| OF11 | GGCGTTCAGATGTCCCGTAT | AAGAGCGAAGAGCACTGGAC |
| OF12 | TCAGGCCGTCACTGGTCTTT | CTGGTGTTCTGGGTGAGAGTC |
| OF13 | TCTGCTTGCAGTTGAACCCA | ACTGCCAGCTTCTTCAAAGGT |
| OF14 | GTGAAGAGAGCACTTGGGGT | AGGGGAAACGACTCACTAGC |
| OF15 | GGCCCACATCAACTTTCCAC | CAGACGGATACAGACAGCACC |
| OF16 | CCATGCTTATGGCCGTTCAG | AGTTTGAACCTCACGGGCTT |
| OF17 | TACAGTAGCGGGTCCTCCAA | TTCCAGCATGGCTGGTTCAT |
| OF18 | TTGCTTTGATGATAAGCCCCG | CAGAAAGCCTCAAGCCTACCA |
| OF19 | TTACCTTGCCCAGAAACCCC | TGAGGGGCAGTCATGTCAAC |
| OF20 | TTTCCACTCTATGTGCCCCTG | CAGTCACACCCTAAGTGCCC |
| OF21 | AAAAGCACTGGCACGTGTGG | ATGGCACAGTTTTCCCGGTAG |
| OF22 | GCCCACCCCCTACTACTCAT | CCCTGCAATTGACTTTCCCG |
| OF23 | CGCGCCTTGTCTCTACAGCTT | TCTGTCTGTCTCTGCTCGCT |
| OF24 | GATCACATTGAGGCCGTGGT | AGCTGATCTGCCCACCAAC |
| OF25 | TAGCTGAGCGGCAAGAAAGAG | AAGGGTGGCCTGCACTTTTG |
| OF26 | GAGACAAGAAGGAGTCCCAGAC | GTTACTGCTTTCACCCTCCCT |
| OF27 | GGTGTTGGAGAAGAGGGTGTC | GGTCGTGCTGAAGAAATCCG |
| OF28 | GGACCATCTGGAGTACCACC | TGCTTCCACTCCTAGGAACTT |
| OF29 | GAGCTATGGAATCAGGGGCT | ACTTCCCAGGCTACAGAAGG |
| OF30 | TTTGCTGAAGAGCCTGCAATG | TTCTCCTACAGTTCCTCACCCA |
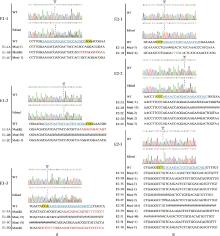
Fig.9
Mutational analysis of monoclonal MSTN knockout cell lines from single vectors①A and B, The mutation status of monoclonal cell clones targeting the MSTN gene exons 1 and 2 obtained through flow cytometric fluorescence sorting, respectively. ②The yellow square represents the PAM sequence; The blue letters are the sgRNA; The red letters are the mutant base; The green letters are the insertion bases. ③WT, Wild-type cells; Edited, Gene-edited cells; MUt, Mutant. ④Different symbols represent different types of mutations, the numbers indicate the quantity of bases inserted or deleted; Mut, Mutant; #, Missing bases; -, Deletion mutation; +, Insertional mutation; &, Multi-base mutations. The same as below"

Table 7
Monoclonal cell line screening statistics"
名称 Name | 突变类型 Type of mutation | 插入/缺失 Insertion/Deletion/bp | 效率 Efficiency/% |
|---|---|---|---|
| E1-1A | 缺失 | 1 | 50.0 (3/6) |
| E1-1B | 多碱基突变 | 0 | |
| E1-1C | 插入 | 1 | |
| E1-2A | 多碱基突变 | 0 | 16.7(3/18) |
| E1-2B | 缺失 | 20 | |
| E1-2C | 插入 | 1 | |
| E1-3A | 多碱基突变 | 0 | 40.0(4/10) |
| E1-3B | 缺失 | 19 | |
| E1-3C | 缺失 | 18 | |
| E1-3D | 多碱基突变 | 0 | |
| E2-1A | 缺失 | 3 | 7.7(2/26) |
| E2-1B | 插入 | 5 | |
| E2-2A | 插入 | 1 | 45.5(5/11) |
| E2-2B | 缺失 | 13 | |
| E2-2C | 缺失 | 2 | |
| E2-2D | 缺失 | 2 | |
| E2-2E | 缺失 | 2 | |
| E2-3A | 插入 | 2 | 57.9(11/19) |
| E2-3B | 插入 | 1 | |
| E2-3C | 缺失 | 1 | |
| E2-3D | 缺失 | 1 | |
| E2-3E | 缺失 | 9 | |
| E2-3F | 缺失 | 2 | |
| E2-3G | 缺失 | 265 | |
| E2-3H | 插入 | 1 | |
| E2-3I | 缺失 | 50 | |
| E2-3J | 插入 | 1 | |
| E2-3K | 缺失 | 1 | |
| E1+3A | 多碱基突变 | 0 | 15.4(2/13) |
| E1+3B | 插入 | 1 | |
| E2+3A | 缺失 | 253 | 55.6(5/9) |
| E2+3B | 多碱基突变 | 0 | |
| E2+3C | 缺失 | 9 | |
| E2+3D | 缺失 | 6 | |
| E2+3E | 缺失 | 253 |
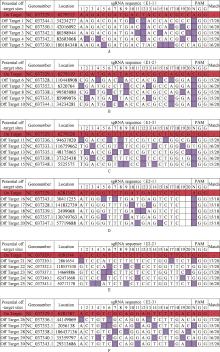
Fig.11
Information on off-target sites of Cas9 vectorA-F, Information about off-target sites for each vector; The red box is the target information corresponding to the vector; The white box indicates that the bases of the potential off-target site are consistent with the target bases, while the purple box indicates that the bases of the both are different"


Fig.12
Off-target sequencing results of Cas9 vector① A, Agarose gel electrophoresis results of PCR products for off-target efficiency detection; B, Sequencing information for off-target sites targeting exons 1 and 2, respectively. ②OF1-OF5 corresponds to five off-target sites of vector E1-1, OF6-OF10 corresponds to five off-target sites of vector E1-2, and so on"

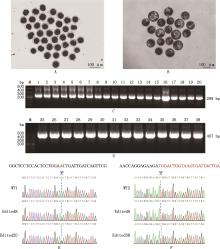
Fig.14
Gene-edition of electroporated embryos① A, Cleavage embryos after electroporation; B, Embryos that have developed to blastocyst stage after electroporation; C and D, Nested PCR amplification results targeting exons 1 (1-20) and 2 (25-38) of MSTN gene,respectively; E and F, Sanger sequencing results of targeting MSTN gene exons 1 and 2 edited embryos,respectively. ②M, DNA Marker Ⅰ; 1-38, Edited blastocyst identifiers"

| [1] | GUO X, LIU C, ZHAO Y, et al. CRISPR ribonucleoprotein-mediated precise editing of multiple genes in porcine fibroblasts[J]. Animals, 2024, 14(4): 650. |
| [2] | MA J, GAO X, LI J Y, et al. Assessing the genetic background and selection signatures of Huaxi cattle using high-density SNP array[J].Animals, 2021, 11(12): 3469. |
| [3] | 厉荣. 我国肉牛产业可持续发展测度及影响因素[J]. 饲料研究, 2024, 47(19): 186-190. |
| LI R. Measurement and influencing factors of sustainable development of China’s beef cattle industry[J]. Feed Research, 2024, 47(19): 186-190. (in Chinese) | |
| [4] | 张天留, 王泽昭, 朱波, 等. 华西牛新品种培育及对我国肉牛育种的启示[J]. 吉林农业大学学报, 2023, 45(4): 385-390. |
| ZHANG T L, WANG Z Z, ZHU B, et al. The cultivation of new breeds of Huaxi cattle and its enlightenment to beef cattle breeding in China[J]. Journal of Jilin Agricultural University, 2023, 45(4): 385-390. (in Chinese) | |
| [5] | MENCHACA A, SANTOS-NETO P C D, MULET A P, et al. CRISPR in livestock: From editing to printing[J]. Theriogenology, 2020, 150: 247-254. |
| [6] | VILLAMIL P R, BEATON B P, KRISHER R L. Gene editing in livestock: Innovations and applications[J]. Animal Reproduction, 2024, 21(3): 20240054. |
| [7] | POPOVA J, BETS V, KOZHEVNIKOVA E. Perspectives in genome-editing techniques for livestock[J]. Animals, 2023, 13(16): 2580. |
| [8] | SHENG H, GUO W Y, ZHANG L L, et al. Proteomic studies on the mechanism of myostatin regulating cattle skeletal muscle development[J]. Frontiers in Genetics, 2021, 12: 752129. |
| [9] | GYEONGMIN G, DONGHYUK K, KYEONGHYUN E, et al. Production of MSTN-mutated cattle without exogenous gene integration using CRISPR-Cas9[J]. Biotechnology Journal, 2021, 17(7): 2100198. |
| [10] | YANG S P, ZHU X X, QU Z X, et al. Production of MSTN knockout porcine cells using adenine base-editing-mediated exon skipping[J]. In Vitro Cellular & Developmental Biology Animal, 2023, 59(4): 241-255. |
| [11] | SONG S Z, HE Z Y, CHENG Y, et al. MSTN modification in goat mediated by TALENs and performance analysis[J]. Hereditas (Beijing), 2022, 44(6): 531-542. |
| [12] | JAKARIA J, WENNY L N A, RIYADI I, et al. Discovery of SNPs and indel 11 bp of the myostatin gene and its association with the double-muscled phenotype in Belgian blue crossbred cattle[J]. Gene, 2021, 784: 145598. |
| [13] | KALDS P, ZHOU S, HUANG S, et al. When less is more: Targeting the Myostatin gene in livestock for augmenting meat production[J].Journal of Agricultural and Food Chemistry, 2023, 71(10): 4216-4227. |
| [14] | 李光鹏, 白春玲, 魏著英, 等. 黄牛Myostatin基因编辑研究[J]. 内蒙古大学学报(自然科学版), 2020, 51(1): 12-32. |
| LI G P, BAI C L, WEI Z Y, et al. Myostatin gene editing study in cattle[J]. Journal of Inner Mongolia University (Natural Science Edition), 2020, 51(1): 12-32. (in Chinese) | |
| [15] | 娜日娜, 冀国尚, 杨梦丽, 等. 华西牛与中国西门塔尔牛肉用性能分析[J]. 中国畜牧杂志, 2025, 61(10): 149-156. |
| NA R N, JI G S, YANG M L, et al. Analysis of beef performance in Huaxi cattle and Chinese Simmental cattle[J]. Chinese Journal of Animal Science, 2025, 61(10): 149-156. (in Chinese) | |
| [16] | 李柯安宁, 杜丽丽, 安炳星, 等. 华西牛胴体及原始分割肉块重量性状遗传参数估计与全基因组关联分析[J]. 畜牧兽医学报, 2023, 54(9): 3664-3676. |
| LI K A N, DU L L, AN B X, et al. Genetic parameter estimation and genome-wide association study for carcass traits and primal cuts weight traits in Huaxi cattle[J]. Acta Veterinaria et Zootechnica Sinica, 2023, 54(9): 3664-3676. (in Chinese) | |
| [17] | MA J, GAO X, LI J Y, et al. Assessing the genetic background and selection signatures of Huaxi cattle using high-density SNP array[J]. Animals, 2021, 11(12): 3469. |
| [18] | KHAN N, LI Z, ALI A, et al. Comprehensive transcriptomic analysis of myostatin-knockout pigs: Insights into muscle growth and lipid metabolism[J]. Transgenic Research, 2025, 34(1): 12. |
| [19] | 朱琳, 谷明娟, 王丽娜,等. Myostatin基因编辑牛骨骼肌组织学结构与转录组分析[J]. 农业生物技术学报, 2022, 30(10): 1903-1912. |
| ZHU L, GU M J, WANG L N, et al. Myostatin gene-edited bovine skeletal muscle histological structure and transcriptome analysis[J]. Journal of Agricultural Biotechnology, 2022, 30(10): 1903-1912. (in Chinese) | |
| [20] | LI H H, WANG G, HAO Z Q, et al. Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer[J]. Scientific Reports, 2016, 6(1): 33675. |
| [21] | ZHANG M, CAI G Y, ZHOU R, et al. Generation of ETV5 knockout pigs with CRISPR/Cas9[J]. Indian Journal of Animal Research, 2021, 55(9): 999-1004. |
| [22] | SUVÁ M, BASTÓN I J, WIEDENMANN A E, et al.Use of an exogenous DNA-free system to generate MSTN-KO calves by CRISPR/Cas9 and SCNT[J]. Reproductive Biology, 2025, 25(3):101050. |
| [23] | REDEL B K, YOON J, REESE E, et al. Novel off-targeting events identified after genome-wide analysis of CRISPR-Cas edited pigs[J]. The CRISPR Journal, 2024, 7(3): 141-149. |
| [24] | RYCZEK N, HRYHOROWICZ M, LIPIŃSKI D, et al. Evaluation of the CRISPR/Cas9 genetic constructs in efficient disruption of porcine genes for xenotransplantation purposes along with an assessment of the off-target mutation formation[J]. Genes (Basel), 2020, 11(6): 713. |
| [25] | MAHESH G, MARTIN E W, AQDAS M, et al. Whole genome sequencing of CRISPR/Cas9-engineered NF-κB reporter mice for validation and variant discovery[J]. Scientific Data, 2024, 11(1): 1225. |
| [26] | HAMZE J G, CAMBRA J M, SERNA S N, et al. Navigating gene editing in porcine embryos: Methods, challenges, and future perspectives[J]. Genomics, 2025, 117(2): 111014. |
| [27] | CHEN J, WANG J, ZHAO H, et al. Molecular breeding of pigs in the genome editing era[J]. Genetics, Selection, Evolution, 2025, 57(1): 12. |
| [28] | PUNETHA M, KUMAR D, SAINI S, et al. Optimising electroporation condition for CRISPR/Cas-mediated knockout in zona-intact buffalo zygotes[J]. Animals, 2023, 14(1): 134. |
| [29] | MIN G G, HYEON E K, HYEOK K D, et al. Generation of double knockout cattle via CRISPR-Cas9 ribonucleoprotein (RNP) electroporation[J]. Journal of Animal Science and Biotechnology, 2023, 14(1): 103. |
| [30] | DU X, QUINN A, MAHONY T, et al. Optimizing genome editing in bovine cells: A comparative study of Cas9 variants and CRISPR delivery methods[J]. Biocatalysis and Agricultural Biotechnology, 2025, 6(5): 103553. |
| [31] | CAMARGO L S A, OWEN J R, VAN EENENNAAM A L, et al. Efficient one-step knockout by electroporation of ribonucleoproteins into zona-intact bovine embryos[J]. Frontiers in Genetics, 2020, 11: 570069. |
| [32] | MAJI D, MIGUELA V, CAMERON A D, et al. Enhancing in vivo electroporation efficiency through hyaluronidase: Insights into plasmid distribution and optimization strategies[J]. Pharmaceutics, 2024, 16(4): 547. |
| [33] | FERNÁNDEZ J P, PETERSEN B, HASSEL P, et al. Comparison between electroporation at different voltage levels and microinjection to generate porcine embryos with multiple xenoantigen knock-outs[J]. International Journal of Molecular Sciences, 2024, 25(22): 11894. |
| [1] | Hanbing ZHANG, Yaping GUO, Jiaqing ZHANG, Qiaoling REN, Junfeng CHEN, Fujiu LIU, Jing WANG, Baosong XING. Research Progress on Gene Editing Technology and Its Application in Breeding of Pigs [J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(1): 1-14. |
| [2] | Linna GAO, Yingying JIANG, Guangjun HUANG, Yue WANG, Qianqian SHI, Huili WANG, Kunlin CHEN. Construction and Functional Study of FGFR1 Gene Knockout Bovine Mammary Epithelial Cell Line [J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(1): 317-332. |
| [3] | WANG Chenyu, CHEN Shihao, BI Yulin, CHEN Guohong, CHANG Guobin, BAI Hao. Gene Editing Technology and Its Application in Poultry Disease-resistant Breeding [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(10): 4765-4775. |
| [4] | JIA Wenfeng, JIANG Xiangxiang, TAO Huili, WANG Anping, WU Zhi, ZHU Shanyuan. Development of a Method for Rapid Construction of Recombinant Duck Enteritis Virus Based on HDR-CRISPR/Cas9 Technology [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(1): 298-309. |
| [5] | PAN Dongxia, WANG Hui, XIONG Benhai, TANG Xiangfang. Research Progress on CRISPR-Cas9 Gene Editing Technology in Cattle and Sheep Production [J]. China Animal Husbandry & Veterinary Medicine, 2024, 51(11): 4880-4889. |
| [6] | TAO Maohai, MENG Ke, RONG Xuan, QIANG Hao, NIE Wei, GUO Chenhao, FENG Dengzhen. Polymorphism of MSTN Gene and Its Association Analysis with Meat Quality Traits in Sheep [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(7): 2766-2776. |
| [7] | FU Yongzhi, MA Xiaojun, WANG Maohan, LI Zhe, CHEN Zhiyuan, QUAN Wei, ZHAO Han, SU Yongmei, WU Jianhua, ZHU Wenjin. Bioinformatics Analysis and Tissue Expression of MSTN Gene in Yangyuan Donkeys [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(8): 2910-2919. |
| [8] | WANG Jing, ZHU Zhe, ZHANG Peng, BI Yanzhen. Site-directed Mutagenesis of Porcine Myostatin Gene Using Single Base Editor [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(8): 2880-2887. |
| [9] | XIE Sihao, GOU Hongchao, BIAN Zhibiao, LI Bin, CAI Rujian, ZANG Yingan, LI Chunling. Effects of MLKL Gene Knockout on Replication of Pseudorabies Virus [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(5): 1934-1941. |
| [10] | XU Jing, YANG Guang, JIANG Meiqi, DING Xiangbin, GUO Yiwen, HU Debao, LI Xin, GUO Hong, ZHANG Linlin. Research Advances on CRISPR/Cas9 Technology in Livestock and Poultry Breeding [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(4): 1374-1383. |
| [11] | LI Xiaojiao, HE Yanhua, ZHU Xinyu, ZOU Xian, LUO Chenglong. Research Progress on Application of CRISPR/Cas9 Technology in Pigs and Chickens [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(12): 4665-4673. |
| [12] | ZHANG Junxia, HE Na, LI Mingming, LYU Cailing, CUI Yajie, LANG Xia, WANG Cailian. Cloning,Bioinformatics and Tissue Expression Analysis of MSTN Gene in Tibetan Sheep [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(11): 4307-4317. |
| [13] | MA Jing, ZHANG Yanyan, JIA Xinyue, MA Xun, WANG Zhengrong, BO Xinwen. Research Progress of CRISPR/Cas9 System in Parasite Genome Editing [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(10): 3713-3725. |
| [14] | CHEN Fuguang, LU Tongyan, LI Shaowu. Bioinformatic Analysis of CRISPR/Cas System in Flavobacterium psychrophilum [J]. China Animal Husbandry & Veterinary Medicine, 2021, 48(6): 1927-1938. |
| [15] | ZHANG Lei, LI Danni, WANG Yuchen, YANG Lichun, HE Jing, SONG Yuxuan. Polymorphism of MSTN Gene and Its Association with Growth Traits in East Friensian Sheep [J]. China Animal Husbandry & Veterinary Medicine, 2021, 48(12): 4537-4544. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||