
China Animal Husbandry & Veterinary Medicine ›› 2026, Vol. 53 ›› Issue (1): 317-332.doi: 10.16431/j.cnki.1671-7236.2026.01.029
• Genetics and Breeding • Previous Articles Next Articles
GAO Linna1,2( ), JIANG Yingying1,3, HUANG Guangjun1,3, WANG Yue1,3, SHI Qianqian1,2, WANG Huili1, CHEN Kunlin1(
), JIANG Yingying1,3, HUANG Guangjun1,3, WANG Yue1,3, SHI Qianqian1,2, WANG Huili1, CHEN Kunlin1( )
)
Received:2025-05-12
Online:2026-01-05
Published:2025-12-26
Contact:
CHEN Kunlin
E-mail:13292751837@163.com;chenkunlin@jaas.ac.cn
CLC Number:
GAO Linna, JIANG Yingying, HUANG Guangjun, WANG Yue, SHI Qianqian, WANG Huili, CHEN Kunlin. Construction and Functional Study of FGFR1 Gene Knockout Bovine Mammary Epithelial Cell Line[J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(1): 317-332.
Table 4
Primer sequences of Real-time quantitative PCR"
基因 Genes | 登录号 Accession No. | 引物序列 Primer sequences (5′→3′) | 片段长度 Product length/bp |
|---|---|---|---|
| GAPDH | NM_001034034 | F:GGCAAGTTCAACGGCACAGT | 129 |
| R: CACCACATACTCAGCACCAGC | |||
| PCNA | NM_001034494 | F: CGGACACGTTGGCACTAGTATT | 138 |
| R: CAGAAGGCATCTTTACTACACAGC | |||
| CyclinD1 | NM_001046273 | F: CCTGGTGAACAAACTCAAGTGG | 140 |
| R: CACAGAGGGCAACGAAGGT | |||
| CyclinE1 | NM_001192776 | F: CGGATACCATGAAAGAGGAAAG | 162 |
| R: CTCTATTACTGTCCCAAGGCTGA | |||
| CSN1S1 | NM_181029 | F: GCGTTACCTGGGTTATCTGGA | 141 |
| R: ATCATAGGTTCTTTCTGTTGGGC | |||
| CSN2 | XM_015471671 | F: CAGCCTGAAGTAATGGGAGTCT | 205 |
| R: ACATGACAGTTGGAGGAAGAGG | |||
| CSN3 | NM_174294 | F: ACCAATACGCTGTGAGAAAGATG | 156 |
| R: GGGTATGGCAGAAATTGATTATT | |||
| ELF5 | NM_001024569 | F: GAAGCCCTGGCGAAGATGT | 118 |
| R: TTCGGTCAACCCGTTCCA | |||
| GLUT1 | NM_174602 | F: CCTGGATGTCCTACCTGAGC | 178 |
| R: CGCACAGTTGCTCCACATAC | |||
| HKⅡ | XM_015473383 | F: AAGATGCTGCCCACCTACG | 123 |
| R: TCGCTTCCCATTCCTCACA | |||
| SCD-1 | NM_173959 | F: CGACCTAAGAGCCGAGAAGC | 162 |
| R: CCAAGGGCATAACGGAATAA |
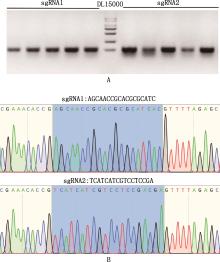
Fig.3
Construction of FGFR1 gene knockout plasmidA, Agarose gel electrophoresis map of PCR amplification product from bacterial liquid; B, FGFR1 gene knockout plasmid sequence alignment map, the blue box selected part is sgRNA alignment successful sequence, and the others are the vector sequence"
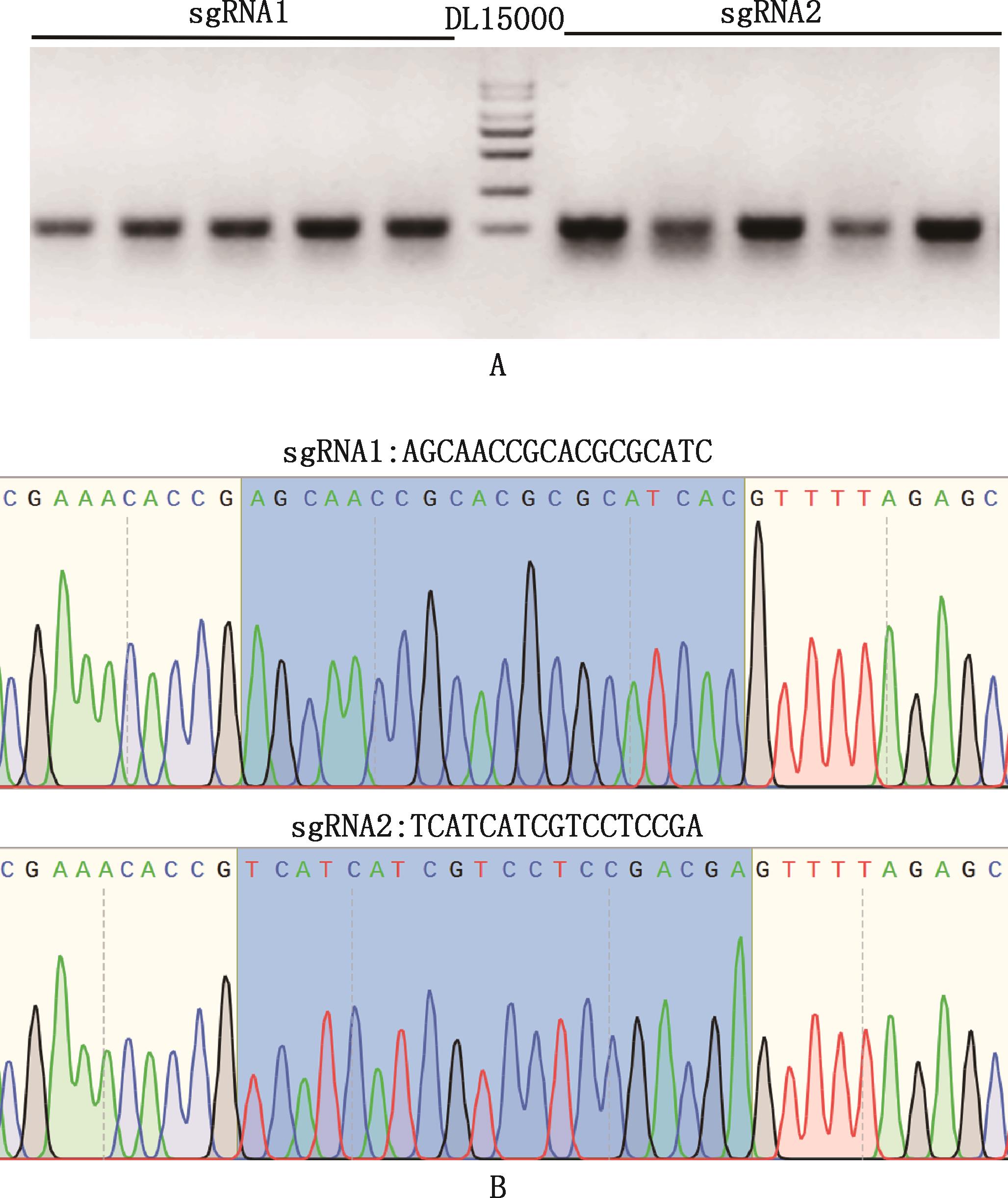
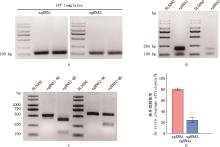
Fig.4
Verification of sgRNA transcription and in vitro cleavage efficiencyA, Agarose gel electrophoresis map of in vitro transcription template; B, Agarose gel electrophoresis map of in vitro transcription of sgRNA, 2 bands represent RNA integrity; C, Agarose gel electrophoresis map of in vitro cleavage of sgRNA, compared with uncut bands (NC group), sgRNA1 and sgRNA2 were cut into bands of different sizes (QG group); D, In vitro cleavage efficiency of sgRNA"

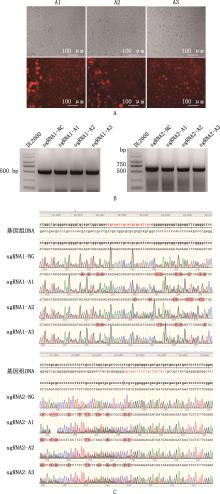
Fig.5
Electrotransfection effect detection of bMECsA, Fluorescence detection of cells after electrotransfection (20×), the upper half show cells under bright field, while the lower half show cells under red fluorescence, A1, A2, and A3 are the three replicates of electroporation; B, Agarose gel electrophoresis map of cells after drug sieve 7 d; C, Sequence alignment results of editing region of cells after drug sieve 7 d, the genomic DNA was NCBI sequence, NC is the untreated cell expansion sequence, sgRNA1/2-A1, sgRNA1/2-A2, and sgRNA1/2-A3 are the cell amplification sequences of knockout group, the red parts are the editing region, and the doublets are multiple editing conditions"

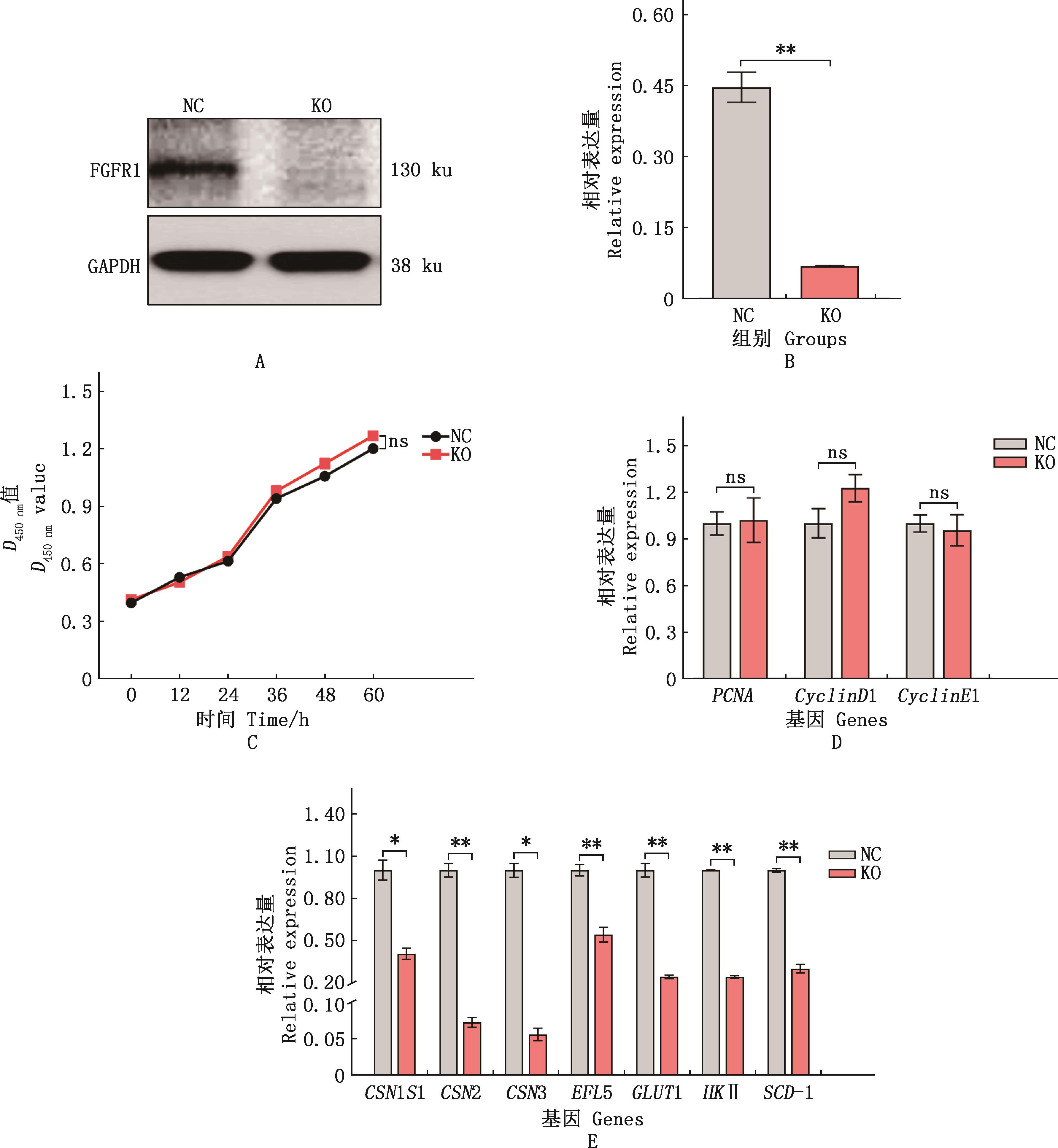
Fig.8
Effects of FGFR1 gene knockout on survival, proliferation and lactation function of bMECs①A, Western blotting analysis of FGFR1 protein expression in monoclonal cell lines; B, ImageJ grayscale analysis results; C, CCK-8 assay for cell viability; D and E, Real-time quantitative PCR analysis of the expression of proliferation and lactation synthesis related genes, respectively. ②*, Significant difference (P<0.05); **, Extremely significant difference (P<0.01); ns, No significant difference (P>0.05)"
| [1] | LIN Q, SERRATORE A, NIU J, et al. Fibroblast growth factor receptor 1 inhibition suppresses pancreatic cancer chemoresistance and chemotherapy-driven aggressiveness[J]. Drug Resistance Updates, 2024, 73: 101064. |
| [2] | FAN S, CHEN Y, WANG W, et al. Pharmacological and biological targeting of FGFR1 in cancer[J]. Current Issues in Molecular Biology, 2024, 46(11): 13131-13150. |
| [3] | 李 娜. FGFR1、p-ERK及p-STAT3在乳腺癌中的表达与临床意义[D]. 郑州:河南大学,2021. |
| LI N. Expression and clinical significance of FGFR1, p-ERK and p-STAT3 in breast cancer [D]. Zhengzhou: Henan University, 2021. (in Chinese) | |
| [4] | 楚阳光, 孙 胜, 邓小强, 等. FGFR1蛋白在ER阳性乳腺癌中的表达情况及其与ER、预后的相关性[J].实用癌症杂志, 2024, 39(2): 208-211. |
| CHU Y G, SUN S, DENG X Q, et al. FGFR1 protein expression in ER-positive breast cancer and its correlation with ER and prognosis[J]. The Practical Journal of Cancer, 2024, 39(2): 208-211. (in Chinese) | |
| [5] | 徐 南, 徐 杰, 李 函, 等. 敲减FGFR1基因表达对婴幼儿血管瘤内皮细胞生物学特性的影响[J]. 中国病理生理杂志, 2019, 35(3): 418-423. |
| XU N, XU J, LI H, et al. Effect of FGFR1 expression knock-down on biological characteristics of infantile hemangioma endothelial cells[J]. Chinese Journal of Pathophysiology, 2019, 35(3): 418-423. (in Chinese) | |
| [6] | 陈美丽, 袁忆航, 杨 晖, 等. 成纤维细胞生长因子受体在胃癌中的研究进展[J]. 中国临床研究, 2024, 37(2): 192-196. |
| CHEN M L, YUAN Y H, YANG H, et al. Research progress of fibroblast growth factor receptor in gastric cancer[J]. Chinese Journal of Clinical Research, 2024, 37(2): 192-196. (in Chinese) | |
| [7] | MÄKINEN N, MEYERSON M. Genomic insights into the mechanisms of FGFR1 dependency in squamous cell lung cancer[J]. The Journal of Clinical Investigation, 2023, 133(21): e174171. |
| [8] | SERVETTO A, KOLLIPARA R, FORMISANO L, et al. Nuclear FGFR1 regulates gene transcription and promotes antiestrogen resistance in ER(+) breast cancer[J]. Clinical Cancer Research, 2021, 27(15): 4379-4396. |
| [9] | CIMMINO F, MONTELLA A, TIRELLI M, et al. FGFR1 is a potential therapeutic target in neuroblastoma[J]. Cancer Cell International, 2022, 22(1): 174. |
| [10] | XIAO H, WANG K, LI D, et al. Evaluation of FGFR1 as a diagnostic biomarker for ovarian cancer using TCGA and GEO datasets [J]. PeerJ, 2021, 9: e10817. |
| [11] | BI Y, ZHENG R, HU J, et al. A novel FGFR1 inhibitor CYY292 suppresses tumor progression, invasion, and metastasis of glioblastoma by inhibiting the Akt/GSK3β/Snail signaling axis[J]. Genes & Diseases, 2024, 11(1): 479-494. |
| [12] | YU T, YANG Y, LIU Y, et al. A FGFR1 inhibitor patent review: Progress since 2010[J]. Expert Opinion on Therapeutic Patents, 2017, 27(4): 439-454. |
| [13] | 王 云, 韩正祥. FGFR1抑制剂Ponatinib对乳腺癌细胞增殖凋亡的影响[J]. 现代肿瘤医学, 2017, 25(8): 1196-1203. |
| WANG Y, HAN Z X, The influence of FGFR 1 inhibitor Ponatinib on celluar proliferation and apoptosis of breast cancer cells[J]. Journal of Modern Oncology, 2017, 25(8): 1196-1203. (in Chinese) | |
| [14] | JEONG W, BAE H, LIM W, et al. The functional effects and mechanisms by which fibroblast growth factor 2 (FGF2) controls bovine mammary epithelial cells: Implications for the development and functionality of the bovine mammary gland[J]. Animal Science, 2017, 95(12): 5365-5377. |
| [15] | 李若诚,王迪铭,邓蒙蒙,等. 奶牛乳腺功能研究的生物材料来源分类及其特征比较[J]. 中国畜牧杂志, 2024, 60(1): 123-130. |
| LI R C, WANG D M, DENG M M, et al. Classification of biological materials sources and comparison of their characteristics for functional studies of mammary glands in dairy cows[J]. Chinese Journal of Animal Science, 2024, 60(1): 123-130. (in Chinese) | |
| [16] | 王若薇, 许曦瑶, 汤晓娜, 等. 结缔组织生长因子体外调控奶牛乳腺上皮细胞生长和泌乳分化[J].畜牧兽医学报, 2024, 55(8): 3446-3459. |
| WANG R W, XU X Y, TANG X N, et al, Connective tissue growth factor regulates the growth and differentiation of cows mammary epithelial cells in vitro [J]. Acta Veterinaria et Zootechnica Sinica, 2024, 55(8): 3446-3459. (in Chinese) | |
| [17] | 刘建霞. 奶牛乳腺上皮细胞体外培养研究进展[J]. 中国奶牛, 2021, 9: 20-24. |
| LIU J X. Advances in in vitro culture of dairy cow mammary epithelial cells[J]. China Dairy Cattle, 2021, 9: 20-24. (in Chinese) | |
| [18] | PACESA M, PELEA O, JINEK M. Past, present, and future of CRISPR genome editing technologies[J]. Cell, 2024, 187(5): 1076-1100. |
| [19] | ZHOU S, KALDS P, LUO Q, et al. Optimized Cas9: sgRNA delivery efficiently generates biallelic MSTN knockout sheep without affecting meat quality[J]. BMC Genomics, 2022, 23(1): 348. |
| [20] | ZHANG J, LIU J, YANG W, et al. Comparison of gene editing efficiencies of CRISPR/Cas9 and TALEN for generation of MSTN knock-out cashmere goats[J]. Theriogenology, 2019, 132: 1-11. |
| [21] | GIM G M, KWON D H, EOM K H, et al. Production of MSTN-mutated cattle without exogenous gene integration using CRISPR-Cas9[J]. Biotechnology Journal, 2022, 17(7): e2100198. |
| [22] | LI R, ZENG W, MA M, et al. Precise editing of myostatin signal peptide by CRISPR/Cas9 increases the muscle mass of Liang Guang Small Spotted pigs[J]. Transgenic Research, 2020, 29(1): 149-163. |
| [23] | MORO L N, VIALE D L, BASTÓN J I, et al. Generation of myostatin edited horse embryos using CRISPR/Cas9 technology and somatic cell nuclear transfer[J]. Scientific Reports, 2020, 10(1): 15587. |
| [24] | ZHOU D, WANG Y, YANG R, et al. The MyoD1 promoted muscle differentiation and generation by activating CCND2 in Guanling cattle[J]. Animals, 2022, 12(19): 2571. |
| [25] | LI M, TANG X, YOU W, et al. HMEJ-mediated site-specific integration of a myostatin inhibitor increases skeletal muscle mass in porcine[J]. Molecular Therapy Nucleic Acids, 2021, 26: 49-62. |
| [26] | ZHOU W, WAN Y, GUO R, et al. Generation of beta-lactoglobulin knock-out goats using CRISPR/Cas9[J]. Public Library of Science ONE, 2017, 12(10): e0186056. |
| [27] | TARA A, SINGH P, GAUTAM D, et al. CRISPR-mediated editing of β-lactoglobulin (BLG) gene in buffalo[J]. Scientific Reports, 2024, 14(1): 14822. |
| [28] | TIAN H, LUO J, ZHANG Z, WU J, et al. CRISPR/Cas9-mediated stearoyl-CoA desaturase 1 (SCD1) deficiency affects fatty acid metabolism in goat mammary epithelial cells[J]. Journal of Agricultural and Food Chemistry, 2018, 66(38): 10041-10052. |
| [29] | WANG L, XUE Z, WANG J, et al. Targeted knockout of Mx in the DF-1 chicken fibroblast cell line impairs immune response against Newcastle disease virus[J]. Poultry Science, 2023, 102(9): 102855. |
| [30] | SHANDILYA U K, SHARMA A, MALLIKARJUNAPPA S, et al. CRISPR-Cas9-mediated knockout of TLR4 modulates Mycobacterium avium ssp. paratuberculosis cell lysate-induced inflammation in bovine mammary epithelial cells[J]. Journal of Dairy Science, 2021, 104(10): 11135-11146. |
| [31] | 丁修虎, 林志平, 赵 芳, 等. 利用CRISPR/Cas9技术制备BLG基因敲除牛乳腺上皮细胞系[J]. 畜牧兽医学报, 2024, 55(10): 4475-4488. |
| DING X H, LIN Z P, ZHAO F, et al. Highly efficient BLG knockout in bovine mammary epithelial cells by using CRISPR/Cas9[J]. Acta Veterinaria et zootechnica Sinica, 2024, 55(10): 4475-4488. (in Chinese) | |
| [32] | TUZON C T, RIGUEUR D, MERRILL A E. Nuclear fibroblast growth factor receptor signaling in skeletal development and disease[J]. Current Osteoporosis Reports, 2019, 17(3): 138-146. |
| [33] | LATKO M, CZYREK A, PORĘBSKA N, et al. Cross-talk between fibroblast growth factor receptors and other cell surface proteins[J]. Cells, 2019, 8(5): 455-455. |
| [34] | SUH J, KIM D H, KIM S J, et al. Nuclear localization of fibroblast growth factor receptor 1 in breast cancer cells interacting with cancer associated fibroblasts[J]. Journal of Cancer Prevention, 2022, 27(1): 68-76. |
| [35] | 胡梦蝶, 张淑君. FGFR结构与功能研究进展[J]. 承德医学院学报, 2022, 39(4): 337-341. |
| HU M D, ZHANG S J. Research progress on the structure and function of FGFR[J]. Journal of Chengde Medical University, 2022, 39(4): 337-341. (in Chinese) | |
| [36] | WEEDEN C E, AH-CANN C, HOLIK A Z, et al. Dual inhibition of BCL-XL and MCL-1 is required to induce tumour regression in lung squamous cell carcinomas sensitive to FGFR inhibition[J]. Oncogene, 2018, 37(32): 4475-4488. |
| [37] | LI J, KONG D, KE Y, et al. Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis[J]. BMC Biology, 2024, 22(1): 6. |
| [38] | MOURIDI S EL, LECROISEY C, TARDY P, et al. Reliable CRISPR/Cas9 genome engineering in Caenorhabditis elegans using a single efficient sgRNA and an easily recognizable phenotype[J]. G3 (Bethesda), 2017, 7(5): 1429-1437. |
| [39] | 唐才智, 任 泂, 高美娇,等. 成纤维细胞生长因子受体2对小肠上皮细胞增殖与分化的影响[J]. 第三军医大学学报, 2018, 40(10): 847-854. |
| TANG C Z, REN J, GAO M J, et al. Effect of fibroblast growth factor receptor 2 on proliferation and differentiation of intestinal epithelial cells in adult mice[J]. Journal of Third Military Medical University, 2018, 40(10): 847-854. (in Chinese) | |
| [40] | 孙 梦, 郑 钢, 吴俊锋, 等. 成纤维细胞生长因子2重组蛋白对鸡原代成肌细胞增殖和分化的影响[J]. 中国家禽, 2024, 46(12): 24-30. |
| SUN M, ZHENG G, WU J F, et al. Effects of recombinant protein of fibroblast growth factor 2 on proliferation and differentiation of chicken primary myoblasts[J]. China Poultry, 2024, 46(12): 24-30. (in Chinese) | |
| [41] | 牛恺文. FGFR1过表达对口咽部鳞癌迁移和侵袭能力的影响[D]. 广州:南方医科大学, 2022. |
| NIU K W. The effects of FGFR1 overexpression on the migration and invasion of oropharyngeal squamous cell carcinoma[D]. Guangzhou: Southern Medical University, 2022. (in Chinese) | |
| [42] | 刘林楠, 范志松, 冯 莉. 抗成纤维细胞生长因子及其受体信号通路药物在肿瘤治疗中应用的研究进展[J]. 中国肿瘤生物治疗杂志, 2023, 30(4): 338-343. |
| LIU L N, FAN Z S, FENG L. Research progress in the application of anti-fibroblast growth factor and its receptor signaling pathway drugs in tumor therapy[J]. Chinese Journal of Cancer Biotherapy, 2023, 30(4): 338-343. (in Chinese) | |
| [43] | YANG Y, LIU C, ZHANG C, et al. Acetate upregulates GPR43 expression and function via PI3K-Akt-SP1 signaling in mammary epithelial cells during milk fat synthesis[J]. Journal of Agricultural and Food Chemistry, 2023, 71(43): 16003-16015. |
| [44] | 邢鹏飞, 王晓雪, 王智慧,等. GPR37L1对奶牛乳腺上皮细胞乳成分合成相关基因表达的影响[J].东北农业大学学报, 2024, 55(4): 22-31. |
| XING P F, WANG X X, WANG Z H, et al. Effects of GPR37L1 on expression of genes related to milk component synthesis in dairy cow mammary epithelial cells[J]. Journal of Northeast Agriultural University, 2024, 55(4): 22-31. (in Chinese) | |
| [45] | POND A C, BIN X, BATTS T, et al. Fibroblast growth factor receptor signaling is essential for normal mammary gland development and stem cell function[J]. Stem Cells, 2013, 31(1): 178-189. |
| [1] | DU Zhiwen, GAO Yuxin, LIU Shuqin, MA Haibin, YANG Lei, SONG Lishuang, BAI Chunling, WEI Zhuying, LI Guangpeng, SU Guanghua. Efficiency Optimization of MSTN Gene Editing in Huaxi Cattle Cells and Embryos [J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(2): 819-836. |
| [2] | ZHANG Hanbing, GUO Yaping, ZHANG Jiaqing, REN Qiaoling, CHEN Junfeng, LIU Fujiu, WANG Jing, XING Baosong. Research Progress on Gene Editing Technology and Its Application in Breeding of Pigs [J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(1): 1-14. |
| [3] | WANG Wei, BI Zhenbin, GU Shanshen, XIAO Yeyi, ZHOU Yajing, WU Shenglong, BAO Wenbin, WANG Haifei. Construction of Porcine TRIM8 Gene Knockout Cell Line Based on CRISPR/Cas9 Technology and Its Regulatory Effect on Porcine Epidemic Diarrhea Virus Replication [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(8): 3790-3799. |
| [4] | WANG Nan, DU Weiwei, WANG Wanjie, WANG Yue, YUAN Maosha, NIE Yuxin, SUN Yaru, LIU Zhiguo, WU Tianwen, MU Yulian. Establishment of ST Cell Lines with WIP1 Gene g.37536832 C>A Mutation Using the CRISPR/Cas9 Gene Editing System [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(5): 1966-1976. |
| [5] | NIE Yuxin, YUAN Maosha, WANG Wanjie, WANG Nan, SUN Yaru, LIU Zhiguo, MU Yulian. Establishment of CRISPR/Cas9-mediated CD71 Gene Editing IPI-2I Cell Line [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(10): 4527-4537. |
| [6] | WANG Chenyu, CHEN Shihao, BI Yulin, CHEN Guohong, CHANG Guobin, BAI Hao. Gene Editing Technology and Its Application in Poultry Disease-resistant Breeding [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(10): 4765-4775. |
| [7] | JIA Wenfeng, JIANG Xiangxiang, TAO Huili, WANG Anping, WU Zhi, ZHU Shanyuan. Development of a Method for Rapid Construction of Recombinant Duck Enteritis Virus Based on HDR-CRISPR/Cas9 Technology [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(1): 298-309. |
| [8] | DONG Zexia, LIN Xin, ZHOU Qilyu, WANG Nan, HUANG Lei, LIU Zhiguo, FENG Zheng, MU Yulian. Construction and PRRSV Infection Characteristic Analysis of CD163 Gene Knockout iPAMs [J]. China Animal Husbandry & Veterinary Medicine, 2024, 51(8): 3471-3483. |
| [9] | ZHANG Xueping, LIU Jiayi, WANG Yanfang, WU Tianwen. Construction of ACTA1 Gene Knockout PEFs Cell Lines by CRISPR/Cas9 Editing System [J]. China Animal Husbandry & Veterinary Medicine, 2024, 51(6): 2273-2284. |
| [10] | DONG Jiao, LU Fan, FANG Xiaomin, CHEN Yuzhe, BAO Wenbin, WANG Haifei. Construction of Porcine KLF4 Gene Knockout Cell Using CRISPR/Cas9 Technology and Its Effect on Cell Viability [J]. China Animal Husbandry & Veterinary Medicine, 2024, 51(3): 893-902. |
| [11] | PAN Dongxia, WANG Hui, XIONG Benhai, TANG Xiangfang. Research Progress on CRISPR-Cas9 Gene Editing Technology in Cattle and Sheep Production [J]. China Animal Husbandry & Veterinary Medicine, 2024, 51(11): 4880-4889. |
| [12] | WANG Hui, FENG Baoliang, XIANG Guangming, HUANG Lei, LIU Zhiguo, LI Kui, MU Yulian. Construction of iPAMs with CD163 Monoallelic Expression and Characterization of Their Mediating PRRSV Infection [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(7): 2617-2628. |
| [13] | ZHANG Jiaxiang, HAN Diangang, SHI Yaling, MAO Xiaoyue, ZHAO Kaiwei, DU Xuan, XIN Jige. Construction of IPEC-J2 Cell Lines with PPARγ Gene Knockout Mediated by CRISPR/Cas9 Technology [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(7): 2670-2677. |
| [14] | SHA Fangfang, FAN Pei, YANG Peichang, ZHANG Lu, LI Jianke. Generation of Mrjp1 Gene Knock-in Mice with Specific Expression in Vascular Smooth Muscle Cells via CRISPR/Cas9 Strategy [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(11): 4392-4402. |
| [15] | MENG Jiejie, SONG Yue, FAN Wenjie, YANG Le, XING Jiayou, WANG Jiang, CHU Beibei, YANG Guoyu, WANG Mengdi. Effect of Toll-like Receptor 7 Gene Knockout on Proliferation of Vesicular Stomatitis Virus [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(6): 2011-2021. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||