
China Animal Husbandry & Veterinary Medicine ›› 2026, Vol. 53 ›› Issue (1): 469-478.doi: 10.16431/j.cnki.1671-7236.2026.01.042
• Basic Veterinary Medicine • Previous Articles Next Articles
LAN Lizhen1,2( ), HUA Baoyu1, FU Mingjun1(
), HUA Baoyu1, FU Mingjun1( )
)
Revised:2025-07-14
Online:2026-01-05
Published:2025-12-26
Contact:
FU Mingjun
E-mail:lizhenlan2021@163.com;fu.mj@163.com
CLC Number:
LAN Lizhen, HUA Baoyu, FU Mingjun. Molecular Characterization and Expression Analysis Under Stress of Citrobacter of Antimicrobial Peptide Cathelicidin from Quasipaa spinosa[J]. China Animal Husbandry & Veterinary Medicine, 2026, 53(1): 469-478.
Table 1
Bioinformmatics analysis software and websites"
软件 Software | 网址 Websites | 功能预测 Function prediction |
|---|---|---|
| ExPASy | https://web.expasy.org/compute_pi/ | 理化性质 |
| NetPhos 3.1 | https://services.healthtech.dtu.dk/services/NetPhos-3.1/ | 磷酸化位点 |
| NetNGlyc 1.0 | https://services.healthtech.dtu.dk/services/NetNGlyc-1.0/ | 糖基化位点 |
| TMHMM 2.0 | https://services.healthtech.dtu.dk/services/TMHMM-2.0/ | 跨膜结构 |
| SignalP 5.0 | https://services.healthtech.dtu.dk/services/SignalP-5.0/ | 信号肽 |
| SMART | https://smart.embl.de/ | 结构域 |
| NovoPro | https://www.novopro.cn/tools/secondary-structure-prediction.html | 二级结构 |
| SWISS-MODEL | https://www.swissmodel.expasy.org/ | 三级结构 |
Table 2
Primer information"
引物名称 Primer name | 引物序列 Primer sequences (5′→3′) | 退火温度 Annealing temperature/℃ | 用途 Application |
|---|---|---|---|
| Cathelicidin-F | ACCATCCCAATCGTCCAC | 55.2 | PCR |
| Cathelicidin-R | CAGGAAAGATTTATTAAGTATTCTGG | ||
| Cathelicidin-qF | AAGACTGCGACCTGGAGAAATG | 60.0 | 实时荧光定量PCR |
| Cathelicidin-qR | AGAACAGCCGTCCCGATGA | ||
| GAPDH-qF | CCTGCCGCCTGGAGAAGC | 60.0 | 实时荧光定量PCR |
| GAPDH-qR | TCAAAGATGGAGGAATGGGTGT |
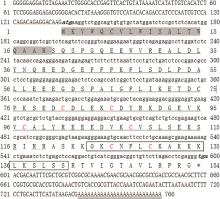
Fig.1
Full length display and deduced amino acid sequence of Cathelicidin gene in Quasipaa spinosaThe italicized and bolded part of the nucleic acid sequence, atg represents the ORF start codon, * and tga represent the ORF stop codon; The gray-shaded region represents the signal peptide of the Cathelicidin gene; The single-underlined segment corresponds to the Cathelicidins domain; Residues highlighted in red indicate conserved cysteine residues; The boxed sequence designates the cleaved mature peptide; And the double-underlined region identifies the poly(A) tail structure"


Fig.6
Multiple sequence analysis of Cathelicidin of Quasipaa spinosa and other speciesIn the figure, residues with extremely high conservation are marked by “*”, while those with moderate conservation are denoted by “:” and “.”, respectively, in descending order of conservation strength; A solid-line box encloses four conserved cysteine residues, and a dashed-line box highlights the Cathelicidin proteolytic cleavage motif (-RR- or -KR-) responsible for releasing the mature peptide; Black dots indicate the Quasipaa spinosa Cathelicidin protein sequence"

| [1] | JEAN S, ARDURA M I. 141-Citrobacter Species[M]. Principles and Practice of Pediatric Infectious Diseases (Sixth Edition).Philadelphia: Elsevier, 2023. |
| [2] | 杨晓伟, 刘于畅, 李毕成, 等. 1株大鲵源葡萄牙柠檬酸杆菌的全基因组测定及毒力基因和耐药基因的分析[J]. 中国兽医学报, 2023, 43(2):291-296. |
| YANG X W, LIU Y C, LI B C, et al. Genome-wide determination of Citrobacter portuguese from a strain of Andrias davidianus and analysis of virulence gene and drug resistance gene[J]. Chinese Journal of Veterinary Science, 2023, 43(2):291-296. (in Chinese) | |
| [3] | 吕美, 郭怡德, 陈耀还,等. 鳄蜥源葡萄牙柠檬酸杆菌和摩氏摩根菌的分离鉴定及药敏分析[J]. 野生动物学报, 2023, 44(2):383-392. |
| LYU M, GUO Y D, CHEN Y H, et al. Isolation, identification and drug susceptibility analysis of Citrobacter staphylococcus and Morganella mosani derived from Shinisaurus crocodilurus [J]. Chinese Journal of Wildlife, 2018, 44(2):383-392. (in Chinese) | |
| [4] | 刘乃瑜, 王至诚, 何宏港,等. 鳖源柠檬酸杆菌属新种的分离鉴定与多位点序列[J]. 水产学报, 2023, 47(3):203-214. |
| LIU N Y, WANG Z C, HE H G, et al. Isolation, identification and multilocus sequence of a new species of Citrobacter derived from Trionychia [J]. Journal of Fisheries of China, 2023, 47(3):203-214. (in Chinese) | |
| [5] | YANG J, TIAN T, XIAO K, et al. Pathogenic infection and immune-related gene expression of Chinese sturgeon (Acipenser sinensis) challenged by Citrobacter freundii [J]. Developmental & Comparative Immunology, 2021, 114:103872. |
| [6] | 程晓云, 郑芊芷, 宋婷婷,等. 棘胸蛙白内障病原鉴定及药敏试验[J]. 浙江农业科学, 2016, 57(7):1141-1143. |
| CHENG X Y, ZHENG Q Z, SONG T T, et al. Pathogen identification and drug susceptibility test of cataracts in Quasipaa spinosa [J]. Journal of Zhejiang Agricultural Sciences, 2016, 57(7):1141-1143. (in Chinese) | |
| [7] | YANG L, GORDO V D, MISHRA A, et al. Synthetic antimicrobial oligomers induce a composition-dependent topological transition in membranes[J]. Journal of the American Chemical Society, 2007, 129(40):12141-12147. |
| [8] | ZANETTI M. Cathelicidins, multifunctional peptides of the innate immunity[J]. Journal of Leukocyte Biology, 2004, 75(1):39-48. |
| [9] | SHINNAR A E, BUTLER K L, PARK H J. Cathelicidin family of antimicrobial peptides: Proteolytic processing and protease resistance[J]. Bioorganic Chemistry, 2003, 31(6):425-436. |
| [10] | BALS R, WILSON J M. Cathelicidins—A family of multifunctional antimicrobial peptides[J]. Cellular and Molecular Life Sciences, 2003, 60(4):711-720. |
| [11] | YANG Y, WU J, LI Q, et al. A non-bactericidal cathelicidin provides prophylactic efficacy against bacterial infection by driving phagocyte influx[J]. eLife, 2022, 11:e72849. |
| [12] | LUO X, OUYANG J, WANG Y, et al. A novel anionic cathelicidin lacking direct antimicrobial activity but with potent anti-inflammatory and wound healing activities from the salamander Tylototriton kweichowensis [J]. Biochimie, 2021, 191:37-50. |
| [13] | MWANGI J, YIN Y, WANG G, et al. The antimicrobial peptide ZY4 combats multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii infection[J]. Proceedings of the National Academy of Sciences of the United States of America, 2019, 116(52):26516-26522. |
| [14] | KASTIN A J. Handbook of Biologically Active Peptides[M].2nd ed. San Diego, CA : Elsevier, 2013. |
| [15] | HAO X, YANG H, WEI L, et al. Amphibian cathelicidin fills the evolutionary gap of cathelicidin in vertebrate[J]. Amino Acids, 2012, 43(2):677-685. |
| [16] | CHEN X, LIU S, FANG J, et al. Peptides isolated from amphibian skin secretions with emphasis on antimicrobial peptides[J]. Toxins, 2022, 14(10):722. |
| [17] | CHAI J, CHEN X, YE T, et al. Characterization and functional analysis of cathelicidin-MH, a novel frog-derived peptide with anti-septicemic properties[J]. eLife, 2021, 10:e64411 . |
| [18] | YANG H, LU B, ZHOU D, et al. Identification of the first cathelicidin gene from skin of Chinese giant salamanders Andrias davidianus with its potent antimicrobial activity[J]. Developmental and Comparative Immunology, 2017, 77:141-149. |
| [19] | 展波, 高媛媛, 林维平,等. 中华大蟾蜍皮肤Cathelicidin家族新型抗菌肽的鉴定及其抗菌活性[J]. 中国中药杂志, 2016, 41(4):630-635. |
| ZHAN B, GAO Y Y, LIN W P, et al. Identification and antibacterial activity of novel antimicrobial peptides from the skin Cathelicidin family of Bufo agargarizans [J]. China Journal of Chinese Materia Medica, 2016, 41(4):630-635. (in Chinese) | |
| [20] | 广慧娟. 脆皮大头蛙Cathelicidins基因克隆、表达及结构与功能分析[D]. 大连:大连理工大学, 2013. |
| GUANG H J. Cloning, expression, structure and function analysis of Cathelicidins gene in Limnonectes fragilis [D]. Dalian: Dalian University of Technology, 2013. (in Chinese) | |
| [21] | TRAVIS S M, ANDERSON N N, FORSYTH W R, et al. Bactericidal activity of mammalian cathelicidin-derived peptides[J]. Infection and Immunity, 2000, 68(5):2748-2755. |
| [22] | 王妍. 黑斑侧褶蛙(Pelophylax nigromaculata)cathelicidin家族抗菌肽的基因克隆、结构与功能研究[D].贵州:贵州师范大学, 2021. |
| WANG Y. Gene cloning, structure and function of cathelicidin family antimicrobial peptides from Pelophylax nigromaculata [D]. Guizhou: Guizhou Normal University, 2021. (in Chinese) | |
| [23] | 马沙, 盛雪晴, 许飘尹, 等. 黄沙鳖抗菌肽cathelicidin基因克隆及表达特征分析[J]. 南方农业学报, 2019, 50(7):1605-1612. |
| MA S, SHENG X Q, XU P Y, et al. Cloning and expression characteristics of cathelicidin gene of Truogx sinensis [J]. Journal of Southern Agriculture, 2019, 50(7):1605-1612. (in Chinese) | |
| [24] | 王梅. 黑眶蟾蜍抗菌肽cathelicidin-DM的基因克隆、组织表达及结构与功能的分析[D] .昆明: 昆明理工大学, 2015. |
| WANG M. Gene cloning, tissue expression and analysis of structure and function of cathelicidin-DM, an antimicrobial peptide of the Black-banded toad[D]. Kunming: Kunming University of Science and Technology, 2015. (in Chinese) | |
| [25] | CRISPE I N. The liver as a lymphoid organ[J]. Annual Review of Immunology, 2009, 27(1):147-163. |
| [26] | JEON K W. International Review of Cell and Molecular Biology[M]. Academic Press: Amsterdam, 2022. |
| [27] | MAO X, YAO R, GUO H, et al. Polysaccharides extract from Vaccaria segetalis seeds inhibits kidney infection by regulating cathelicidin expression[J]. Journal of Ethnopharmacology, 2021, 267:113505. |
| [28] | HAFEEZ A BIN, JIANG X, BERGEN P J, et al. Antimicrobial peptides: An update on classifications and databases[J]. International Journal of Molecular Sciences, 2021, 22(21): 11691. |
| [29] | KEISARI Y, OFEK I. The Biology and Pathology of Innate Immunity Mechanisms[M]. Boston, MA:Experimental Medicine and Biology, 2002, 479:203-218. |
| [30] | 姜玉松, 张文琪, 樊汶樵,等. 棘腹蛙皮肤抗菌肽Cathelicidin-Pb的克隆及表达分析[J]. 淡水渔业, 2015, 45(4):26-30. |
| JIANG Y S, ZHANG W Q, FAN W Q, et al. Cloning and expression analysis of Cathelicidin-Pb in the skin of Quasipaa boulengeri [J]. Freshwater Fisheries, 2015, 45(4):26-30. (in Chinese) | |
| [31] | 薛巧, 赵战勤, 刘会胜,等. 弗氏柠檬酸杆菌对动物和人致病性研究进展[J]. 动物医学进展, 2015, 36(7):81-85. |
| XUE Q, ZHAO Z Q, LIU H S, et al. Research progress on the pathogenicity of Citrobacter freundii to animals and humans[J]. Progress in Veterinary Medicine, 2015, 36(7): 81-85. (in Chinese) | |
| [32] | 胡霭臻, 俞艳, 周超,等. 棘胸蛙柠檬酸杆菌的分子鉴定及防治技术[J]. 浙江农业科学, 2018, 59(11):2101-2105. |
| HU A Z, YU Y, ZHOU C, et al. Molecular identification and control technology of Citrobacter sp. in Quasipaa spinosa [J]. Journal of Zhejiang Agricultural Sciences, 2018, 59(11): 2101-2105. (in Chinese) | |
| [33] | BROEKMAN D C, GUÐMUNDSSON G H, MAIER V H. Differential regulation of cathelicidin in salmon and cod[J]. Fish & Shellfish Immunology, 2013, 35(2):532-538. |
| [34] | 宋婷婷. 棘胸蛙规模化养殖常见疾病病原的鉴定及疫苗研究[D] 金华:浙江师范大学, 2015. |
| SONG T T. Identification of pathogens causing common diseases in large-scale breeding of Quasipaa spinosa and vaccine research[D]. Jinhua: Zhejiang Normal University, 2015. (in Chinese) |
| [1] | LIANG Lina, ZHI Yan, WU Yungerzele, HU Ge. Research Progress on Properties, Functions and Optimization Strategies of Antimicrobial Peptides [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(7): 3359-3371. |
| [2] | LI Xiaoyin, LIU Meng, WU Xinxue, LUO Fan, GAO Yanhua. Effects of β-defensin yTAP on Intestinal Morphology and Microbiota Diversity in Mice [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(5): 2379-2391. |
| [3] | HAN Jinhui, ZHAI Pei, LYU Wenping. Effect of Single Amino Acid Substitution on Bioactivity of Heterozygous Peptide KL-21 [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(5): 2400-2408. |
| [4] | GUO Jiajia, NIE Jing, QU Jiu, Danzengzhandu, LI Xiaowei, JIANG Mingfeng, LIU Yili. Mechanism of Antibacterial Action of Antimicrobial Peptides and Their Application in Livestock and Poultry [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(2): 934-945. |
| [5] | DENG Lihuan, YAO Yingying, LI Haiying, WU Yingping, LU Qingqing, WU Run, MA Yue, WANG Zening. Effects of Antimicrobial Peptides and Glucose Oxidase on Growth Performance, Meat Quality,Immune Function and Antioxidant Capacity of Suckling Pigeons [J]. China Animal Husbandry & Veterinary Medicine, 2025, 52(11): 5192-5206. |
| [6] | LIAN Kaiqi, WANG Yuhang, ZENG Yajing, ZHOU Lingling, ZHANG Yuanchen, ZHANG Mingliang, ZHANG Gaiping, WANG Shuangshan. Antimicrobial Mechanism of Antimicrobial Peptide LL-1 Against Klebsiella pneumoniae ATCC 700603 [J]. China Animal Husbandry & Veterinary Medicine, 2024, 51(5): 2237-2244. |
| [7] | BUAYISHAM Kuerban, WANG Yixin, YANG Chengxi, GENG Zijian, XU Yingqing, LUO Gang. Recombinant Expression of Cecropin A and Lysozyme Heteropeptide in Pichia pastoris and Its Activity Analysis [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(12): 5032-5042. |
| [8] | FENG Yanan, YANG Na, MAO Ruoyu, HAO Ya, MA Xuanxuan, TENG Da, WANG Jianhua. Determination of Antimicrobial Peptide NZ2114 in Rat Feed by High Performance Liquid Chromatography [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(11): 4463-4472. |
| [9] | HUANG Jingsheng, ZHU Shuxin, KANG Weichao, DENG Zhijie, YANG Yunmei, LIU Chengzhi, YI Dandan, HE Jiakang, LIANG Zhengmin. Study on Antibacterial Peptide BNBD5 Regulating the Innate Immune Response of Lung Against Actinobacillus pleuropneumoniae [J]. China Animal Husbandry & Veterinary Medicine, 2023, 50(10): 4261-4269. |
| [10] | DENG Li, LI Jie, LIU Baoguo, HANG Bolin. Molecular Design and Antimicrobial Activity Analysis of Antimicrobial Peptides Derived from Bovine Hemoglobin β-subunit [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(9): 3589-3598. |
| [11] | YU Xiaojun, LI Dandan, LI Xuewu, WANG Qun, WANG Guang, WANG Lihua. Effects of Antibacterial Peptide on the Immune Function and Intestinal Tissue Morphology of Growing Female Minks [J]. China Animal Husbandry & Veterinary Medicine, 2022, 49(2): 443-453. |
| [12] | ZHANG Xin, PAN Chenhao, ZHAO Ruili, JIANG Xuan, JIN Tianming, ZHAO Yang, MA Jifei, YU Enyuan, LI Liuan, ZHANG Zhenzhou, LI Guixia, LI Ruizhong. Inhibitory Effect of Antimicrobial Peptide Temprine-La(FS) on Biofilm Formation of Streptococcus suis Type 2 [J]. China Animal Husbandry & Veterinary Medicine, 2021, 48(1): 324-337. |
| [13] | ZHAI Pei, HAN Jinhui, PAN Xiaoyu. Design and Activity Identification of Hybrid Peptide Based on Cecropin A and Mastoparan [J]. China Animal Husbandry & Veterinary Medicine, 2020, 47(9): 3006-3013. |
| [14] | ZHANG Qingjuan, MA Xuanxuan, TENG Da, YANG Na, WANG Xiumin, MAO Ruoyu, HAO Ya, SHAN Yuxue, LIU He, FAN Huan, WANG Jianhua. Study on Antibacterial Effect of Antimicrobial Peptide NZ2114 Against Streptococcus dysgalactiae Isolated from Bovine Mastitis [J]. China Animal Husbandry & Veterinary Medicine, 2020, 47(8): 2603-2614. |
| [15] | SHAN Yuxue, YANG Na, TENG Da, WANG Xiumin, MAO Ruoyu, HAO Ya, MA Xuanxuan, FAN Huan, WANG Jianhua. The Disinfection Effect of Antimicrobial Peptide NZ2114 on Streptococcus agalactiae Isolated from Bovine Mastitis and Its Biofilm [J]. China Animal Husbandry & Veterinary Medicine, 2020, 47(7): 2284-2294. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||